BEST is a free phylogenetics package of programs to estimate the posterior distribution of species trees using multilocus molecular data that accounts for deep coalescence but not for other issues such as horizontal transfer. It is intended to implement the Bayesian hierarchical model proposed by Liang Liu and Dennis Pearl and further developed in collaboration with Scott Edwards. Details of the Bayesian hierarchical model are available in Liang's Thesis (pdf format).
Estimating the posterior distribution of species trees involves two consecutive Markov Chain Monte Carlo (MCMC) procedures. The first MCMC is performed in BEST part 1 (a modified version of MrBayes 3.1.2) in which a new function is added to approximate the joint probability of gene trees. The output gene trees (multilocus) then form the input file of the second MCMC program BEST part 2. Beginning with version 1.6, the weights for the importance sampling component (details in Liang's Thesis) are also calulated within BEST.
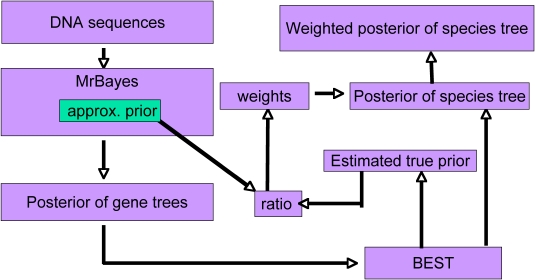
The Bayesian Hierarchical model has been applied to several mutilocus datasets including finch, yeast, and macaques (NEXUS files). Interesting results from these analyses can be found in this technical report and in publications by Liu and Pearl (Systematic Biology, 56:504-514, 2007) and by Edwards, Liu, and Pearl (PNAS, 104:5936-5941, 2007).
Version 1 Program Packages
Thanks to Patricia Brito for preparing the first draft of these basic instructions on getting started with version 1.6. This file contains an example of how to run the two programs and very brief instructions about compiling.
Source Code:
BEST part 1 (version 1.6 - a modification of MrBayes 3.1.2)
The above version of MrBayes includes Liang Liu's code to approximate the joint prior of gene trees given the species tree. MrBayes is the popular Bayesian phylogenetics program by John Huelsenbeck, Fredrik Ronquist, Paul van der Mark, Bret Larget, and Donald SImon. See the MrBayes site for information about its use.
BEST part 2 (version 1.6)
These files allow for the post-processing of the approximate gene tree posterior distribution provided by the revised MrBayes (version allows multiple alleles per species). Program options allow for population sizes to be estimated or to be integrated out of the species tree distribution.
| Version # | Date | Comment |
|---|---|---|
| 1.1 - 1.4 | August 2004 | single allele case only |
| 1.5 | October 2006 | multiple allele case added |
| 1.6 | June 2007 | importance sampling now done within BEST part 2 outfile from part 1 formatted for use in BEST part 2 |
Executables:
Windows version (version 1.6)
Executable for the Windows version of BEST part 2 allowing for multiple alleles per species.
Macintosh version part 1 (version 1.6)
Macintosh version part 2 (version 1.6)
Macintosh executables for the version of BEST parts 1 and 2 allowing for multiple alleles per species. (Note - these are not Finder applications - they must be run from within your terminal program.)
Version 1 Applications
(note: example NEXUS files for version 1.6 uploaded April, 2008)
| NEXUS file | type | # of species (alleles) | # of loci | Reference |
|---|---|---|---|---|
| Finch | single allele | 3 + outgroup | 30 | Jennings, W.B. and Edwards, S.V. Evolution 59:2033-2047, 2005. |
| Yeast | single allele | 8 | 106 | Rokas, A., Williams, B.L., King, N. and Carroll, S.B. Nature 425:798-804, 2003. |
| Yeast | multiple allele | 6 (22) | 4 | Liti, G., Barton, D.B.H., and Louis, E.J. Genetics 174:839-850, 2006. |
| Macaques | single allele | 19 | 4 | Tosi, A.J., Morales, J.C., and Melnick, D.J. Evolution 57:1419-1435, 2003. |
| Primate | single allele | 4 | 53 | Chen, F.C. and Li, W.H. American Journal of Human Genetics 68:444-456, 2001. |


